Staphylococcus aureus is a ubiquitous, versatile and highly adaptive pathogen that colonizes the epidermis and mucosa of the pharynx, Alimentary canal, gastrointestinal system, and anterior slits [1]. Staphylococcus aureus is a causative agent that causes a variety of infections in man and animals, with serious consequences for health [2]. Host specialization, loss resistance and virulence genes have important public health implications [3,2]. Staphylococcus aureus is the more causative agent of the Staphylococci and the causative factor of disease for a wide range of diseases ranging from superficial skin abscess, food poisoning, and life-threatening diseases such as bacteremia and necrotic pneumonia in children, acute respiratory infection Endocarditis [4]. The development of numerous putative virulence factors, as well as the existence of resistance genes like mecA gene, VanA gene, Staphylococcal exotoxins, and other elements that promote disease onset, immune evasion, and the development of antibiotic resistance in host tissues, are what contribute to the disease's severity. When S. aureus first appeared in the mid-1940s, it was described as bacteria. S. aureus produces a hydrolyzing enzyme called penicillinase, which makes it resistant to penicillin [5].
Since then, numerous strains of S. aureus that are resistant to penicillin have been discovered in cases of bacteremia in England and the United States. Resistance isolates were first solely isolated from individuals and healthcare personnel, giving rise to the name hospital-related penicillin-resistant S. aureus. However, resistant bacteria that did not exhibit risk factors associated with hospital isolates were subsequently isolated from members of the public [6]. Methicillin, a semi-synthetic analogue of penicillin, was introduced into clinics as the preferred drug for treating S. aureus infectious illness, that led to a surge in penicillin resistance from the late 1940s through the early 1960s [7].
Methicillin resistance in S. aureus, on the other hand, was discovered within a year of it being presented as that of the preferred strategic therapy for the treatment of S. aureus. The achievement of a genomic archipelago comprising the methicillin-resistant determinant mecA causes MRSA. Methicillin-resistant bacteria have obtained worldwide notoriety as the most widely known cause of man and community-related infections since their discovery during the 1960s in the United Kingdom. As a result, the therapeutic value of several important antibiotics is reduced, and hospitalization time is extended [8].
Classification
Staphylococcus aureus is a facultative anaerobe that is non-motile and non-spore forming, gram-positive, and catalase as well as coagulase positive. Staphylococci are categorized as follows: in irregular grape-like clusters or sometimes singly or in pairs, Typical colonies on blood agar plates with 5% blood are yellow to golden yellow and hemolytic [9,10].
Methicillin-Resistant S. Aureus
MRSA was first identified in 1961, a year after the introduction of methicillin. MRSA strains have since spread throughout hospitals and around the world. MRSA incidence in large US hospital services increased from 4% in the 1980s to 50% in the late 1990s, based on the National Nosocomial Infections Surveillance System. Methicillin-resistant strains record for up to 80% of all Staphylococcus aureus strains in some hospitals. Although MRSA is identified as a significant pathogen in hospitals, MRSA has emerged outside of care settings and spread in the community in recent decades. MRSA strains found in the community have also been linked to healthcare-related infections.
MRSA genetic analysis revealed that the mecA gene contains a motile genetic component called the staphylococcal cassette chromosome mec (SCCmec). There are currently nine various kinds of SCCmec elements identified (types I-VIII and VT). While all nine mobile cassettes contain the entire mecA gene, they each have a slightly various genetic materials term that can affect expression of genes, virulence, and other properties. Bacteria can be described at the strain level, which identifies variations in DNA between bacteria from the same genus [11,12].
The Panton-Valentine leukocidin (PVL) gene is a significant genetic variation that is carried by many isolates of MRSA. More proof is required to support the idea that this gene, which is present in several MRSA strains associated with communities, increases virulence by killing leukocytes and inducing inflammation. The arginine catabolic transportable element is the other significant genetic variation seen in MRSA strains. The USA300 MRSA strain is the only one with this feature. Because ACME contains numerous arc genes that increase bacterial fitness, perhaps leading to strains that are harder to treat and control, it is significant.
Epidemiology
MRSA was first thought to be a hospital virus that primarily affected persons who had contact with healthcare institutions or other risk factors like substance addiction [11]. Healthcare-associated MRSA or healthcare-acquired MRSA are other names for this form of MRSA (HA-MRSA). The frequency of HAMRSA is highest in Latin America, South America, Europe, and Asia, where it has spread to multiple strains [8]. It is important to highlight that the incidence of HA-MRSA varies significantly by region, which researchers believe is because eradication programs and eradication efforts have been successful.
When MRSA infection was first discovered in healthy individuals lacking access to health care or other risk factors, isolation of MRSA in medical institutions started to shift in the 1990s [11]. New MRSA strains have emerged, according to further investigation. Due to their prevalence in healthy communities, these strains were eventually referred to as community-related or community-acquired MRSA [11]. The genetic make-up, related risk factors, and prevalence of the CA-MRSA and HAMRSA strains differ significantly from one another. The type of SCCmec that is incorporated into the genome accounts for the majority of the genetic variations between HA-MRSA and CA-MRSA. The mecA gene is present on bigger SCCmec types in HA-MRSA strains I, II, or III.
The risk factors connected to HA-MRSA or CAMRSA colonization or infection are entirely dissimilar. The major risk factors for acquiring HA-MRSA are exposure to public hospitals and a variety of health difficulties [11]. Also, elderly people are more likely to be admitted to hospitals with HA-MRSA genotypes. Pneumonia, bacteremia, and invasive infections are the most typical HA-MRSA infection kinds [13]. Contrarily, CA-MRSA infection is typically found in healthy individuals who have not recently been exposed to healthcare [14].
Additionally, CA-MRSA strains appear to be more aggressive and easily spread, and they largely damage the skin and infections of soft tissue [15]. Military personnel, inmates, children in care settings, and members of the armed forces have all been found to be at higher risk for CA-MRSA [16]. It is crucial to remember that CA-MRSA strains have spread throughout healthcare facilities and are also prevalent in the general population.
Transmission and Virulence Factors
Because of its proclivity to colonize a wide range of animals, the S. aureus bacterium can quickly have transmitted from one organisms to another. Humans to animals and back again. Zoonotic transmission of staphylococcal infections occurs. It can be spread from animals to people by bites and scrapes, working with organs or bones from diseased animals, and contact with sick animals or carriers that cause skin sores [17].
Due to virulence factors such as hyluronidase, proteases, lipases, nucleases that help in penetration of bacteria; a blood clot that converts fibrinogen to fibrinogen; agglomerating agents fibrinogen-binding proteins; protein A that avoids darkening and phagocytosis; Panton-Valentine leucocidin lead to organs damage; Due to virulence factors such as hyluronidase, protease, lipase, nuclease that help in penetration of bacteria; coagulase enzyme that fibrinogen is converted to fibrin; clumping factors; fibrinogen-binding proteins, protein A that avoids opsonization; PVL is to blame for necrotic changes, enterotoxin; Alpha and beta haemolysins, exfoliative toxin (A, B), toxic shock syndrome toxin-1, and S. aureus exhibit a wide spectrum of illnesses [18,19].
S. aureus has the capsule, which is a viscous layer of polysaccharide that inhibits the ingestion process by leukocytes, and 90% of S. aureus isolates possess the capsule, and the wall of Gram-positive bacteria consists of Peplidoglycan and Lipotechoic acid, which have a role in the mechanism of adhesion cells that covalently associate with specialized proteins to induce infection in S. aureus [20]. Also, some S. aureus species possess the so-called toxic shock syndrome toxin, which is a protein molecule encoded by chromosome genes and this toxin leads to the manifestation of the clinical pattern of toxic shock syndrome. Toxins produced by S. aureus have the potential to cause cellular damage (Figure 1). S. aureus toxins play a significant part in the progression and progression of the infection in the DFI patient.
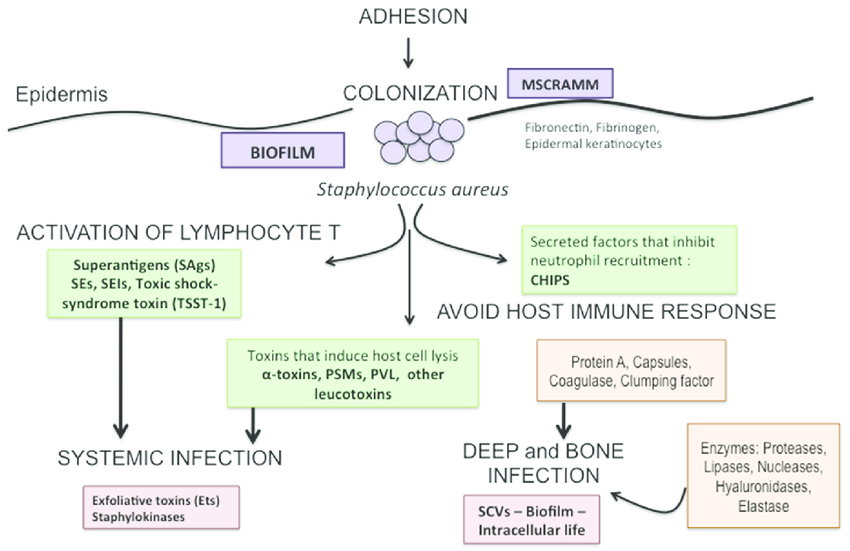
Figure 1: Infection process and virulence factors of Staphylococcus aureus
Treatment of Antibiotic Resistance
The first-line therapy for staphylococcal infections is beta-lactam medication. Alternative medications are available, however they have drawbacks such tissue penetration and decreased effectiveness of vancomycin drug; cost of tigecycline drug, daptomycin, linezolid; and toxicity for rifampin [21]. Because of the selective pressure put on it by antimicrobials, S. aureus has the opportunity to develop drug resistance more quickly, which is a problem that affects the entire world [22]. Resistance is caused by transduction, transformation, and conjugation, which are all chromosome or plasmid-related processes [23].
Tolerance to beta-lactam antibiotics is mediated by the blaZ gene in the organism. The mecA gene, which encodes a phosphate-binding protein with potentially enhanced binding capability or a novel PBP, may be responsible for the resistance to methicillin and other -lactam antimicrobial agents. Some S. aureus (BORSA) strains are resistant to the antibiotic boderline oxyacillin. These organisms create super-lactamases and resist oxacillin when mecA or mecC are absent [23].
Aminoglycosidal resistance may develop as a result of mutations in the genes that control the ribosome, which may cause architectural alterations in proteins that reduce the antibiotic's ability to bind and enhance the antibiotic's impact. The aacA-aphD gene's resistance to antibiotics may also be brought on by decreased absorption of antibiotic or antibiotic-modifying cations by cellular enzymes like aminoglycoside acetyltransferases catalyzed by a bi-functional protein expressed by the aacA-aphD gene [17]. The tetK and tetL genes found on the plasmid are responsible for tetracycline resistance. Active flux is governed by these genes. The tetO or tetM genes mediate ribosome protection.
Antibiotic resistance is a growing concern, and it is caused by the subtherapeutic use of these drugs to enhance growth and prevent sickness, which results in the creation of resistant microbes. Worldwide reports of instances associated with MRSA have been made. Spreading of resistant strains to vulnerable populations is facilitated by population mixing caused by international travel, touch with an infected person, or consumption of contaminated food. Tracking the spread of these antibiotic-resistant isolates is crucial for developing mitigation measures [24,25].



